A gene is a unit of DNA that contains the information to specify the synthesis of a single polypeptide chain or functional RNA(for eg. tRNA). A vast majority of genes in organisms carry information to build protein molecules. Among them, the mRNA molecules are the RNA copies of such protein-coding genes.

During RNA synthesis, the four base languages of DNA containing A, G, C, and T is simply copied i.e. transcribed into four base language of RNA, which is identical except Uracil(U) replaces T. Similar process yields the tRNAs and rRNAs by tRNA and rRNA genes. Some non-protein coding RNA helps to regulate the transcription of protein-coding genes.
During the process of transcription of DNA, one DNA strand act as a template (the antisense strand) which determines the order in which ribonucleotide triphosphate(rNTP) monomers are linked together to form a complementary RNA chain. The other strand is called the coding strand(sense strand). It has the same sequence as mRNA except that U replaces T.
Initiation of RNA synthesis:
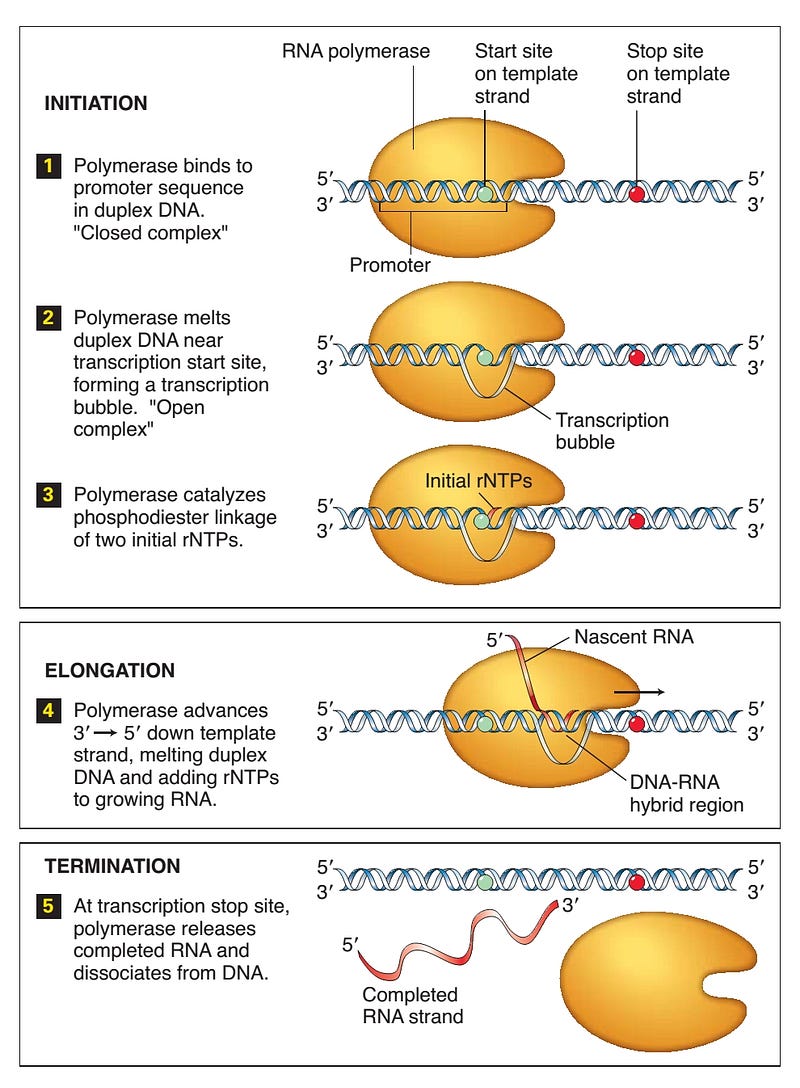
The process of transcription starts when the RNA polymerase binds to the promoter region in the duplex DNA. The RNA polymerase with the help of initiation factors recognizes and binds to a specific sequence of double-stranded DNA called the promoter.
In general terms, a promoter is a sequence in DNA that is especially recognized by proteins called transcription factors.
In prokaryotes, the promoter consists of two short sequences at -10 and -35 positions upstream from the transcription start site.

Downstream: Direction in which DNA strand(template) is transcribed.
Upstream: Opposite to downstream.
Positions of downstream nucleotides are denoted by + sign while of upstream by -ve sign from the initiation point.
- The sequence at -10 is called the Pribnow box or the TATA box or the -10 element and usually consists of the six nucleotides TATAAT. The Pribnow box is absolutely essential to start transcription in prokaryotes.
- The other sequence at –35 (the -35 element) usually consists of the six nucleotides TTGACA. Its presence allows a very high transcription rate.
The promoter sequences are not equally conserved throughout the evolution and are asymmetrical, which means the RNA polymerase gets directional information about the starting point for the transcription from the promoter.
Bacteria contain a single type of RNA polymerase (DNA dependent- RNA Polymerase) that transcribes all the essential RNAs, namely mRNAs, tRNAs, rRNAs, and even smaller microRNAs. The bacterial RNA polymerase essentially functions in two distinct forms.

The first one is the core enzyme, and the second holo-enzyme. The core enzyme is tetrameric in a structure consisting of 2α, ß, ß’ (two alpha, one ß, and one ß’ subunit). This core enzyme can copy the DNA to complementary mRNA, but may not recognize the promoter site. The recognition of the promoter site by the bacterial RNA polymerase is the function of its specialized subunit called the sigma(σ) factor. The core enzyme with its sigma(σ) subunit is now called the holo-enzyme.
Sigma Factor:
Bacteria contain a variety of sigma factors, which recognize different promoter sequences. This implies that it is the sigma factor that determines the transcription of a certain gene. All the bacteria possess a primary sigma factor that transcribes the essential(housekeeping genes). There are other alternative sigma factors whose concentration in cells may increase or decrease depending on the various stress and stimuli or the environmental conditions. The symbiotic E. coli which lives in the gut of other organisms contain seven sigma factors.
The initiation of transcription involves a short unwinding of the DNA double helix to form a complex which is generally accomplished in two steps:
- First RNA polymerase binds to the promoter sequence to form a closed complex. which is relatively weak and can be easily distorted.
- The double-stranded DNA is melted by Polymerase near the transcription start site, forming a transcription bubble i.e. the open complex which is much stronger.
NOTE: Transcription bubble is the unwinding of 12–14 nucleotide pairs in DNA.
At the initiation point, the initiator nucleotide- rNTP monomers are linked together to form a complementary RNA chain. Bases in the template DNA strand pair with the complementary rNTPs which are then joined in a polymerization reaction catalyzed by RNA polymerase.
The reaction involves a nucleophilic attack by the 3′ Oxygen in the growing RNA chain on the α-phosphate of the next nucleotide precursor to being added which results in the formation of a phosphodiester bond and the release of pyrophosphate (PPi). As a consequence of this mechanism, RNA molecules are always synthesized in the 5’ — ›3′ direction and the point is called the active site. After the first nucleotide forms phosphodiester bonds in the complex, the sigma factor is released which now pushes the remaining core polymerase into the elongation mode.
Elongation:
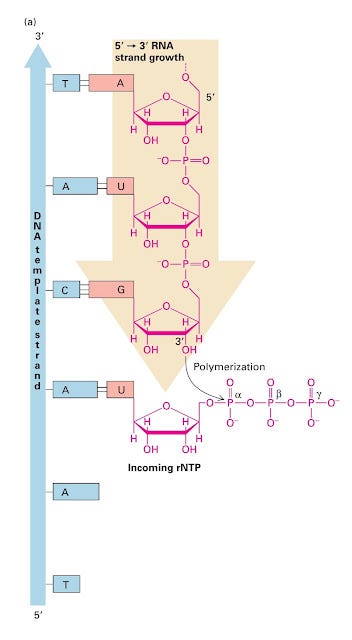
Elongation is the function of core enzyme. RNA polymerase moves along the template DNA, opening the double-stranded DNA in front and its direction of movement and guiding the strands back together, to form intermediate base pairs between the fresh nucleotides, the nascent RNA, and the DNA template strand.
One ribonucleotide at a time is added by the polymerase to the 3′ end of the growing(nascent) RNA chain. During the strand elongation, the enzyme maintains a melted region of approximately 14–20 base pairs in the transcription bubble. Approximately, 8 nucleotides at the 3′ end of the growing RNA strand remain base-paired to the template DNA strand in the transcription bubble. The elongation complex comprising RNA polymerase, template DNA, and the nascent DNA strand is extraordinarily stable.
For instance, a eukaryotic RNA polymerase transcribes the longest known mammalian gene, containing about two million base pairs without dissociating from the DNA template or releasing the nascent RNA. The transcription occurs at a rate of about 1000–2000 nucleotides per minute at 37ºC. So the elongation complex must remain intact for more than 24 hours to ensure continuous synthesis of pre-mRNA from a long gene.
Energetics of Polymerisation Reaction:
The energetics of the polymerization reaction strongly favor the addition of ribonucleotides to the growing RNA chain because of high energy bond between the α and ß phosphates of rNTP monomers is replaced by the lower energy phosphodiester bond between the nucleotides. The equilibrium for the reaction is driven further toward chain elongation by the pyrophosphate, an enzyme that catalyzes the cleavage of the released PPi into two molecules of organic phosphate. Similar to the DNA strands, the newly synthesized RNA strand also has opposite 5’ — ›3′ bi-directionality.
Termination:
Just as transcription is initiated at a specific point in the chromosome, it also terminates when a particular nucleotide sequence is reached. The termination, however, however, depends on the specific sequences in the mRNA which mark the end of the transcribed gene and triggers the release of mRNA from the complex.
Transcription termination is, however, classified into two types.
Intrinsic Termination:
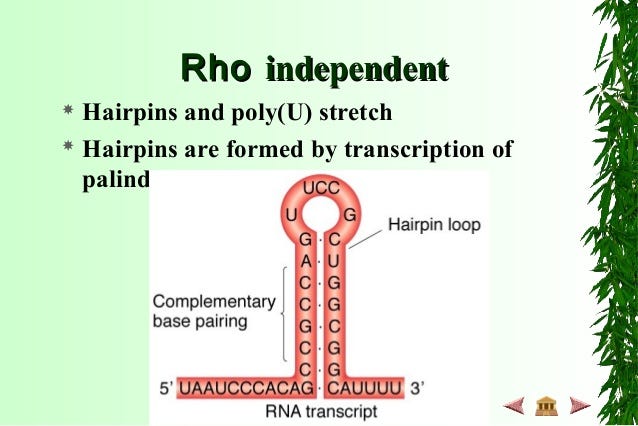
It occurs when the termination sequences typically fold into a hairpin loop that causes the RNA polymerase to release the completed RNA chain without requiring additional factors. The characteristic structure of intrinsic terminators are:
a) A G-C rich stem and loop.
b) a sequence of about 4–6 Uracil residues in the RNA.
Intrinsic termination is sometimes termed as Rho-Independent termination.
Factor(Rho)-Dependent Termination:
Several factors like Rho, Tau, and NusA are important in transcription termination in prokaryotic transcription. In roughly half of the cases, a ring-shaped protein called Rho(ρ) assists with termination. RNA Polymerase reaches a Rho-utilization (rut) site and transcribes this sequence. Rho recognizes this sequence and binds to it. This binding does not occur if a ribosome is bound to the rut site and is translating it. Using energy from ATP hydrolysis, Rho moves along the RNA transcript towards RNA Polymerase. While RNA Polymerase pauses at a Rho-termination site in the DNA, Rho catches up to RNA Polymerase and uses its RNA-DNA helicase activity to cause RNA Polymerase to dissociate. These events result in the termination of transcription.
References:
LODISH, H. F. (2016). Molecular cell biology. New York, W.H. Freeman, and Co.
http://www.scfbio-iitd.res.in/tutorial/promoter.html
https://www.cliffsnotes.com/study-guides/biology/biochemistry-ii/rna-and-transcription/transcription-in-prokaryotes

Post a Comment